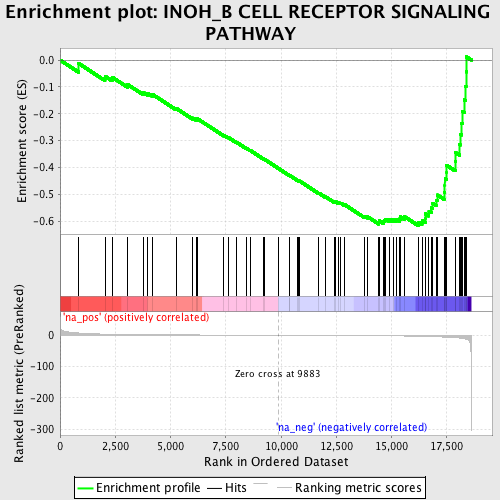
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_B CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.6180952 |
| Normalized Enrichment Score (NES) | -1.8010826 |
| Nominal p-value | 0.009398496 |
| FDR q-value | 0.33424333 |
| FWER p-Value | 0.996 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TRAF4 | 836 | 7.311 | -0.0120 | No | ||
| 2 | CRK | 2032 | 3.614 | -0.0601 | No | ||
| 3 | MYD88 | 2356 | 3.069 | -0.0637 | No | ||
| 4 | BCAR1 | 3058 | 2.335 | -0.0909 | No | ||
| 5 | PRKCZ | 3761 | 1.851 | -0.1204 | No | ||
| 6 | SRC | 3975 | 1.750 | -0.1240 | No | ||
| 7 | SHC1 | 4178 | 1.653 | -0.1274 | No | ||
| 8 | FRS3 | 5251 | 1.241 | -0.1796 | No | ||
| 9 | PRKCE | 6003 | 1.007 | -0.2155 | No | ||
| 10 | HRAS | 6193 | 0.946 | -0.2215 | No | ||
| 11 | PRKCQ | 6222 | 0.935 | -0.2187 | No | ||
| 12 | GRAP2 | 7413 | 0.614 | -0.2801 | No | ||
| 13 | PIK3R2 | 7604 | 0.563 | -0.2878 | No | ||
| 14 | GRB10 | 7966 | 0.468 | -0.3052 | No | ||
| 15 | PRKCI | 8434 | 0.353 | -0.3288 | No | ||
| 16 | TRAF3 | 8599 | 0.313 | -0.3362 | No | ||
| 17 | NRAS | 9203 | 0.170 | -0.3679 | No | ||
| 18 | SHC3 | 9261 | 0.155 | -0.3703 | No | ||
| 19 | DOK2 | 9866 | 0.004 | -0.4028 | No | ||
| 20 | IRS1 | 10369 | -0.122 | -0.4294 | No | ||
| 21 | LAT | 10401 | -0.130 | -0.4304 | No | ||
| 22 | GRB7 | 10735 | -0.219 | -0.4474 | No | ||
| 23 | CRADD | 10804 | -0.235 | -0.4500 | No | ||
| 24 | PRKCH | 10831 | -0.243 | -0.4503 | No | ||
| 25 | LCP2 | 11709 | -0.483 | -0.4954 | No | ||
| 26 | TRAF1 | 11997 | -0.558 | -0.5084 | No | ||
| 27 | MAP2K2 | 12431 | -0.690 | -0.5286 | No | ||
| 28 | MAP2K1 | 12474 | -0.704 | -0.5277 | No | ||
| 29 | GAB2 | 12608 | -0.747 | -0.5315 | No | ||
| 30 | SOS1 | 12695 | -0.778 | -0.5326 | No | ||
| 31 | PIK3R1 | 12882 | -0.833 | -0.5388 | No | ||
| 32 | CBL | 13764 | -1.164 | -0.5811 | No | ||
| 33 | NFKBIB | 13906 | -1.223 | -0.5832 | No | ||
| 34 | ARAF | 14425 | -1.487 | -0.6044 | Yes | ||
| 35 | GRB14 | 14444 | -1.499 | -0.5986 | Yes | ||
| 36 | ITPR3 | 14626 | -1.593 | -0.6011 | Yes | ||
| 37 | FADD | 14667 | -1.621 | -0.5960 | Yes | ||
| 38 | IRS4 | 14736 | -1.655 | -0.5922 | Yes | ||
| 39 | TRAF5 | 14897 | -1.743 | -0.5929 | Yes | ||
| 40 | TRAF2 | 15073 | -1.866 | -0.5939 | Yes | ||
| 41 | LCK | 15211 | -1.955 | -0.5925 | Yes | ||
| 42 | FYN | 15378 | -2.088 | -0.5920 | Yes | ||
| 43 | ITPR1 | 15399 | -2.111 | -0.5835 | Yes | ||
| 44 | PIK3CA | 15572 | -2.276 | -0.5825 | Yes | ||
| 45 | RAF1 | 16233 | -3.067 | -0.6042 | Yes | ||
| 46 | PRKCD | 16381 | -3.304 | -0.5972 | Yes | ||
| 47 | SH2D2A | 16533 | -3.627 | -0.5890 | Yes | ||
| 48 | CD19 | 16540 | -3.644 | -0.5728 | Yes | ||
| 49 | CRKL | 16694 | -3.936 | -0.5633 | Yes | ||
| 50 | PIK3CD | 16811 | -4.218 | -0.5505 | Yes | ||
| 51 | CD79A | 16865 | -4.314 | -0.5339 | Yes | ||
| 52 | SOS2 | 17023 | -4.655 | -0.5213 | Yes | ||
| 53 | BLNK | 17078 | -4.805 | -0.5025 | Yes | ||
| 54 | NFKB1 | 17383 | -5.733 | -0.4930 | Yes | ||
| 55 | RELA | 17416 | -5.813 | -0.4685 | Yes | ||
| 56 | DOK1 | 17420 | -5.829 | -0.4423 | Yes | ||
| 57 | GRB2 | 17489 | -6.003 | -0.4188 | Yes | ||
| 58 | MAPK3 | 17495 | -6.022 | -0.3919 | Yes | ||
| 59 | NFKBIA | 17884 | -7.579 | -0.3786 | Yes | ||
| 60 | NFKBIE | 17895 | -7.630 | -0.3447 | Yes | ||
| 61 | TRAF6 | 18077 | -8.765 | -0.3148 | Yes | ||
| 62 | PRKCA | 18108 | -9.028 | -0.2756 | Yes | ||
| 63 | CD79B | 18176 | -9.666 | -0.2356 | Yes | ||
| 64 | GAB1 | 18219 | -10.102 | -0.1922 | Yes | ||
| 65 | VAV1 | 18300 | -10.774 | -0.1479 | Yes | ||
| 66 | SH2B3 | 18355 | -11.634 | -0.0982 | Yes | ||
| 67 | SYK | 18377 | -12.306 | -0.0438 | Yes | ||
| 68 | KRAS | 18389 | -12.525 | 0.0122 | Yes |
